Planar Imaging#
Overview#
The planar imaging module analyzes phantom images taken with the kV or MV imager in 2D. The following phantoms are supported:
Leeds TOR 18
Standard Imaging QC-3
Standard Imaging QC-kV
Las Vegas
Elekta Las Vegas
Doselab MC2 MV
Doselab MC2 kV
SNC kV
SNC MV
PTW EPID QC
Features:
Automatic phantom localization - Set up your phantom any way you like; automatic positioning, angle, and inversion correction mean you can set up how you like, nor will setup variations give you headache.
High and low contrast determination - Analyze both low and high contrast ROIs. Set thresholds as you see fit.
Feature table#
Feature/Phantom |
Can be inverted? |
SSD setting |
Auto-centering |
Auto-rotation |
Doselab MC2 (MV) |
No |
Manual |
Yes |
Semi (+/-5 from 0) |
Doselab MC2 (kV) |
No |
Manual |
Yes |
Semi (+/-5 from 0) |
Las Vegas |
No |
Manual |
Yes |
No (0) |
Elekta Las Vegas |
No |
Manual |
Yes |
No (0) |
Leeds TOR |
Yes |
Manual |
Yes |
Yes |
PTW EPID QC |
No |
Manual |
Yes |
No (0) |
SNC MV |
No |
Manual |
Yes |
No (45) |
SNC MV (12510) |
No |
Manual |
Yes |
No (45) |
SNC kV |
No |
Manual |
Yes |
No (135) |
SI QC-3 (MV) |
No |
Manual |
Yes |
Semi (+/-5 from 45/135) |
SI QC kV |
No |
Manual |
Yes |
Semi (+/-5 from 45/135) |
IBA Primus A |
No |
Manual |
Yes (+/-2cm) |
Semi (+/-5 from 0,90,270) |
Typical module usage#
The following snippets can be used with any of the phantoms in this module; they all have the same or very similar methods. We will use the LeedsTOR for the example, but plug in any phantom from this module.
Running the Demo#
To run the demo of any phantom, create a script or start an interpreter session and input:
from pylinac import LeedsTOR # or LasVegas, DoselabMC2kV, etc
LeedsTOR.run_demo()
A figure showing the phantom, low contrast plot, and RMTF will be generated:
(Source code, png, hires.png, pdf)
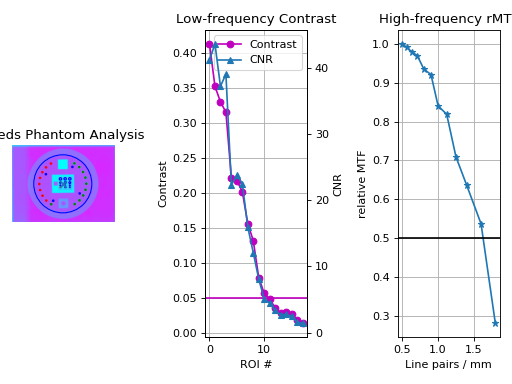
Typical Use#
Import the class:
from pylinac import (
LeedsTOR,
) # or whatever phantom you like from the planar imaging module
The minimum needed to get going is to:
Load image – Load the planar image as you would any other class: by passing the path directly to the constructor:
leeds = LeedsTOR("my/leeds.dcm")
Alternatively, a URL can be passed:
leeds = LeedsTOR.from_url("http://myserver.com/leeds")
You may also use the demo image:
leeds = LeedsTOR.from_demo_image()
Analyze the images – Analyze the image using the
analyze()method. The low and high contrast thresholds can be specified:leeds.analyze(low_contrast_threshold=0.01, high_contrast_threshold=0.5)
Additionally, you may specify the SSD of the phantom. By default, SAD and 5cm up from SID are searched:
leeds.analyze(..., ssd=1400)
View the results – The results of analysis can be viewed with the
plot_analyzed_image()method.leeds.plot_analyzed_image()
(
Source code,png,hires.png,pdf)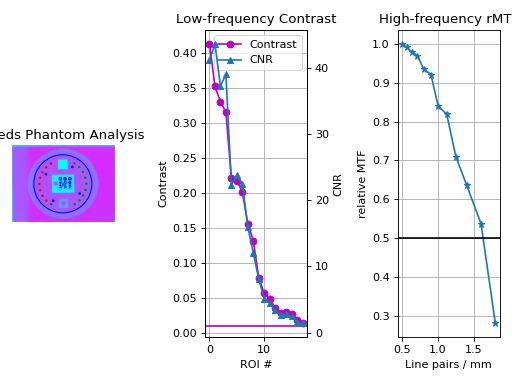
Note that each subimage can be turned on or off.
# don't show the low contrast plot leeds.plot_analyzed_image(low_contrast=False)
The figure can also be saved:
leeds.save_analyzed_image("myprofile.png")
A PDF report can also be generated:
leeds.publish_pdf("leeds_october16.pdf")
Leeds TOR Phantom#
The Leeds phantom is used to measure image quality metrics for the kV imager of a linac. It contains both high and low contrast ROIs.
Note
There are two phantom analysis routines. The LeedsTOR class is for newer phantoms that have a red ring
on the outside. Older Leeds phantoms may have a blue label containing the serial number and model on the back. Use the
LeedsTORBlue class for these phantoms. The difference is small ROI location shifts.
Image Acquisition#
You can acquire the images any way you like. Just ensure that the phantom is not touching a field edge. It is also recommended by the manufacturer to rotate the phantom to a non-cardinal angle so that pixel aliasing does not occur for the high-contrast line pairs.
Algorithm#
Leeds phantom analysis is straightforward: find the phantom in the image, then sample ROIs at the appropriate locations.
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any size kV imager.
The phantom can be at any distance.
The phantom can be at any angle.
The phantom can be flipped either way.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must not be touching or close to any image edges.
The blades should be fully or mostly open to correctly invert the image. This may not result in a complete failure, but you may have to force-invert the analysis if this case isn’t true (i.e.
myleeds.analyze(invert=True)).The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
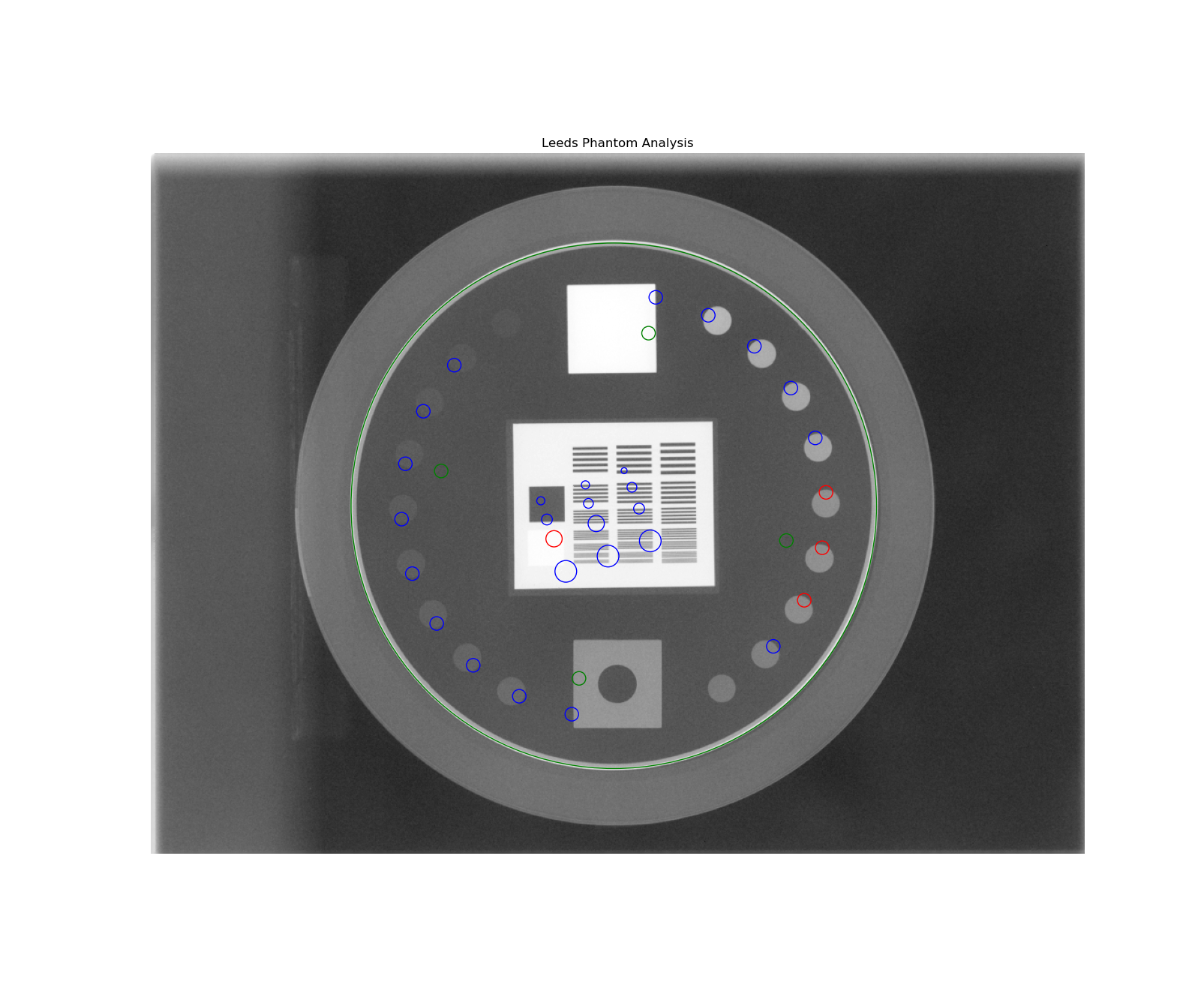
Determine phantom location – The Leeds phantom is found by performing a Canny edge detection algorithm to the image. The thin structures found are sifted by finding appropriately-sized ROIs. This may include the outer phantom edge and the metal ring just inside. The average central position of the circular ROIs is set as the phantom center.
Determine phantom angle – To find the rotational angle of the phantom, a similar process is employed, but square-like features are searched for in the edge detection image. Because there are two square areas, the ROI with the highest attenuation (lead) is chosen. The angle between the phantom center and the lead square center is set as the angle.
Determine rotation direction – The phantom might be placed upside down. To keep analysis consistent, a circular profile is sampled at the radius of the low contrast ROIs starting at the lead square. Peaks are searched for on each semicircle. The side with the most peaks is the side with the higher contrast ROIs. Analysis is always done counter-clockwise. If the ROIs happen to be clockwise, the image is flipped left-right and angle/center inverted.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Calculate high contrast – Again, because the phantom position and angle are known, offsets are applied to sample the high contrast line pair regions. For each sample, the relative MTF is calculated. See Modulation Transfer Function.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
Troubleshooting#
If you’re having trouble getting the Leeds phantom analysis to work, first check out the Troubleshooting section. If the issue is not listed there, then it may be one of the issues below.
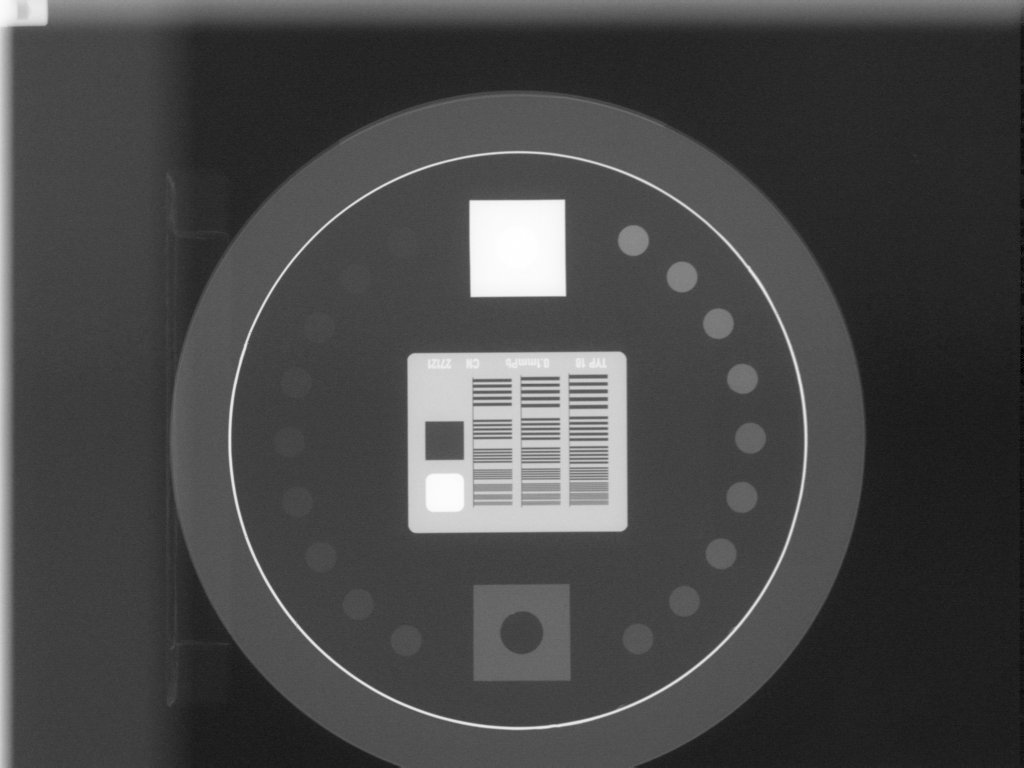
The most common reason for failing is having the phantom near an image edge. The resulting error is usually that the phantom angle cannot be determined. For example, this image would throw an error:
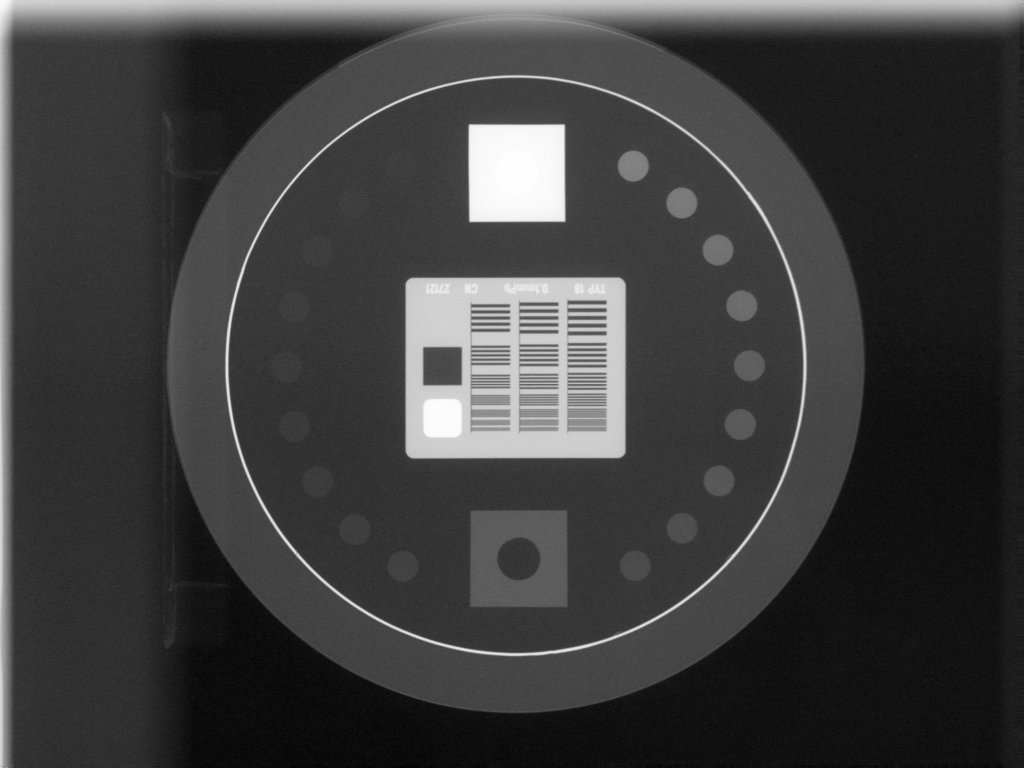
The below image also fails. Technically, the phantom is in the image, but the top blade skews the pixel values such that the phantom edge cannot be properly found at the top. This fails to identify the true phantom edge, causing the angle to also not be found:
Another problem is that the image may have a non-uniform background. This can cause pylinac’s automatic inversion correction to incorrectly invert the image. For example, this image falsely inverts:
When analyzed, the angle is 180 degrees opposite the lead square, causing the ROIs to be
flipped 180 degrees. To correct this problem, pass invert=True to analyze().
This will force pylinac to invert the image the opposite way and correctly identify the lead square.
Another common problem is an offset analysis, as shown below:
This is caused by a wrong inversion.
Note
If the image flash is dark, then the image inversion is very likely wrong.
Again, pass invert=True to the analyze method. This is the same image but with invert=True:

PTW EPID QC Phantom#
The PTW EPID QC phantom is an MV imaging quality assurance phantom and has high and low contrast regions, just as the Leeds phantom, but with different geometric configurations.
Image Acquisition#
The EPID QC phantom appears to have a specific setup as recommended by the manufacturer. The phantom should have the high-contrast line pairs at the top of the image and low contrast at the bottom. The rotation is not automatically determined, so you should take care when setting up the phantom to be well-positioned.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
The images can be acquired with the phantom at any SSD.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must be at 0 degrees.
The phantom must not be touching any image edges.
The phantom should have the high-contrast linen pair regions toward the gantry stand/top.
The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
Determine phantom location – A Canny edge search is performed on the image. Connected edges that are semi-round and angled are thought to possibly be the phantom. Of the ROIs, the one with the longest axis is said to be the phantom edge. The center of the bounding box of the ROI is set as the phantom center.
Determine phantom radius – The major axis length of the ROI determined above serves as the phantom radius.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Calculate high contrast – Again, because the phantom position and angle are known, offsets are applied to sample the high contrast line pair regions. For each sample, the relative MTF is calculated. See Modulation Transfer Function.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
Standard Imaging QC-3 Phantom#
The Standard Imaging phantom is an MV imaging quality assurance phantom and has high and low contrast regions, just as the Leeds phantom, but with different geometric configurations.
Image Acquisition#
The Standard Imaging phantom has a specific setup as recommended by the manufacturer. The phantom should be angled 45 degrees, with the “1” pointed toward the gantry stand and centered along the CAX. For best results when using pylinac, open the jaws to fully cover the EPID, or at least give 1-2cm flash around the phantom edges.
Warning
If using the acrylic jig that comes with the phantom, place a spacer of at least a few mm between the jig and the phantom. E.g. a slice of foam on each angled edge. This is because the edge detection of the phantom may fail at certain energies with the phantom abutted to the acrylic jig.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
The images can be acquired with the phantom at any SSD.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must be at 45 degrees.
The phantom must not be touching any image edges.
The phantom should have the “1” pointing toward the gantry stand.
The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
Determine phantom location – A Canny edge search is performed on the image. Connected edges that are semi-round and angled are thought to possibly be the phantom. Of the ROIs, the one with the longest axis is said to be the phantom edge. The center of the bounding box of the ROI is set as the phantom center.
Determine phantom radius and angle – The major axis length of the ROI determined above serves as the phantom radius. The orientation of the edge ROI serves as the phantom angle.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Calculate high contrast – Again, because the phantom position and angle are known, offsets are applied to sample the high contrast line pair regions. For each sample, the relative MTF is calculated. See Modulation Transfer Function.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
Troubleshooting#
If you’re having issues with the StandardImaging class, make sure you have correctly positioned the phantom as per
the manufacturer’s instructions (also see Image Acquisition). One issue that may arise is incorrect
inversion. If the jaws are closed tightly around the phantom, the automatic inversion correction may falsely
invert the image, just as for the Leeds phantom. If you have an image that looks inverted or just plain weird, add invert=True
to analyze(). If this doesn’t help, reshoot the phantom with the jaws open.
Las Vegas Phantom#
The Las Vegas phantom is for MV image quality testing and includes low contrast regions of varying contrast and size.
There is also a ElektaLasVegas class that is very similar. This section covers
both styles.
Image Acquisition#
The Las Vegas phantom has a recommended position as stated on the phantom. Pylinac will however account for shifts and inversions. Best practices for the Las Vegas phantom:
Keep the phantom from a couch edge or any rails.
The field edge should be >=5mm from the phantom edge, preferably 10+mm.
The orientation should have the largest “holes” towards the right side although this can be accounted for as an
analyzeparameter.The angle should be as close to 0 as possible, given above, although this can be accounted for as an
analyzeparameter.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must not be touching any image edges.
The phantom should be at a cardinal angle (0, 90, 180, or 270 degrees) relative to the EPID.
The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
Determine phantom location – A Canny edge search is performed on the image. Connected edges that are semi-round and angled are thought to possibly be the phantom. Of the ROIs, the one with the longest axis is said to be the phantom edge. The center of the bounding box of the ROI is set as the phantom center.
Determine phantom radius and angle – The major axis length of the ROI determined above serves as the phantom radius. The orientation of the edge ROI serves as the phantom angle.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
Doselab MC2 MV & kV#
The Doselab MC2 phantom is for both kV & MV image quality testing and includes low and high contrast regions of varying contrast. There are two high contrast sections, one intended for kV and one for MV.
Image Acquisition#
The Doselab phantom has a recommended position as stated on the phantom. Pylinac will however account for shifts and inversions. Best practices for the Doselab phantom:
Keep the phantom away from a couch edge or any rails.
Center the phantom along the CAX.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must not be touching any image edges.
The phantom should be at 45 degrees relative to the EPID.
The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
Determine phantom location – A canny edge search is performed on the image. Connected edges that are semi-round and angled are thought to possibly be the phantom. Of the ROIs, the one with the longest axis is said to be the phantom edge. The center of the bounding box of the ROI is set as the phantom center.
Determine phantom radius and angle – The major axis length of the ROI determined above serves as the phantom radius. The orientation of the edge ROI serves as the phantom angle.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
SNC MV & kV#
The SNC MV and kV phantoms are for kV & MV image quality testing and includes low and high contrast regions of varying contrast.
Image Acquisition#
The SNC phantoms typically use the angled setup jig. Best practices for the Doselab phantom:
Keep the phantom away from a couch edge or any rails.
Center the phantom along the CAX.
Use the angled setup jig.
For the MV phantom, have the longer side point inferiorly (i.e. away from the stand).
For the kV phantom, have the longer side point superiorly (i.e. toward the stand).
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must not be touching any image edges.
The phantom should be at 45 degrees relative to the EPID.
The phantom should be centered near the CAX (<1-2cm).
Pre-Analysis
Determine phantom location – A canny edge search is performed on the image. Connected edges that are semi-round and angled are thought to possibly be the phantom. Of the ROIs, the one with the longest axis is said to be the phantom edge. The center of the bounding box of the ROI is set as the phantom center.
Determine phantom radius – The major axis length of the ROI determined above serves as the phantom radius.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
IBA Primus A#
The IBA Primus A phantom is used for kV image analysis and includes low and high contrast regions of varying contrast.
Image Acquisition#
Lay the phantom on the couch with the wedge step circle facing the top/gun and high-res square facing the bottom/target.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom must not be touching any image edges.
The phantom should be at 0, 90, or 270 +/-5 degrees relative to the EPID where 0 is facing the gun.
The dynamic wedge steps should be facing the gun side; the high-resolution square should be facing the target side.
The phantom should be centered near the CAX (<2cm).
Pre-Analysis
Determine phantom location – A Canny edge search is performed on the image. The ROI that approximates the size of the central crosshair of the phantom and is nearly at the center of the image is used as the phantom center location
Determine phantom radius – The size of the above crosshair ROI is used as the basis for the phantom radius.
Fine-tune phantom angle – The phantom angle is assumed to be around 0 (wedge steps facing gun), but fine-tuning is performed so that sensitive ROIs like MTF can be had with high accuracy. This is performed by taking a circular profile about the phantom at the radius of the wedge steps. The two areas of highest gradient will be at the first and last wedge steps. The center between these two points is the angle at which the phantom is “pointing” and will be used as the updated angle.
Warning
If the gradients cannot be found or if the determined angle is >5 degrees (caused by bad inversion, e.g.) a warning will be printed to the console and a default of 0 will be used.
Analysis
Calculate low contrast – Because the phantom center and angle are known, the angles to the ROIs can also be known. From here, the contrast can be known; see Contrast.
Calculate high contrast – Again, because the phantom position and angle are known, offsets are applied to sample the high contrast line pair regions. For each sample, the relative MTF is calculated. See Modulation Transfer Function.
Post-Analysis
Determine passing low and high contrast ROIs – For each low and high contrast region, the determined value is compared to the threshold. The plot colors correspond to the pass/fail status.
Standard Imaging FC-2#
The FC-2 phantom is for testing light/radiation coincidence.
Image Acquisition#
The FC-2 phantom should be placed on the couch at 100cm SSD.
Keep the phantom away from a couch edge or any rails.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom should be at a cardinal angle (0, 90, 180, or 270 degrees) relative to the EPID.
The phantom should be centered near the CAX (<1cm).
The phantom should be +/- 1cm from 100cm SSD.
Pre-Analysis
Determine BB set to use – There are two sets of BBs, one for 10x10cm and another for 15x15cm. To get the maximum accuracy, the larger set is used if a 15x15cm field is irradiated. The field size is determined and if it’s >14cm then the algorithm will look for the larger set. Otherwise, it will look for the smaller 4.
Analysis
Get BB centroid – Once the BB set is chosen, image windows look for the BBs in a 1x1cm square. Once it finds them, the centroid of all 4 BBs is calculated.
Determine field center – The field size is measured along the center of the image in the inplane and crossplane direction. A 5mm strip is averaged and used to reduce noise.
Post-Analysis
Comparing centroids – The irradiated field centroid is compared to the EPID/image center as well as the the BB centroid. The field size is also reported.
Customizing behavior#
The BB window as well as the expected BB positions, and field strip size can be overridden like so:
from pylinac import StandardImagingFC2
class MySIFC2(StandardImagingFC2):
bb_sampling_box_size_mm = (
20 # look at a 20x20mm window for the BB at the expected position
)
# change the 10x10 BB expected positions. This is in mm relative to the CAX.
bb_positions_10x10 = {
"TL": [-30, -30],
"BL": [-30, 30],
"TR": [30, -30],
"BR": [30, 30],
}
bb_positions_15x15 = ... # same as above
field_strip_width_mm = 10 # 10mm strip in x and y to determine field size
# use as normal
fc2 = MySIFC2(...)
Doselab RLf#
New in version 3.15.
The Doselab RLf is for testing light/radiation coincidence. See also DoselabRLf.
Image Acquisition#
The RLf phantom should be placed on the couch at 100cm SSD.
Keep the phantom away from a couch edge or any rails.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom should be at a cardinal angle (0, 90, 180, or 270 degrees) relative to the EPID.
The phantom should be centered near the CAX (<2mm).
Analysis
Get BB centroid – An image window looks for each BB on the inner side of each edge. After finding the BBs, the centroid is calculated.
Note
The inner 10x10 BBs are always used regardless of the field size. This is because the BB detection is more robust when the BBs are away from a field edge. This also means that 10x10 analysis is slightly less robust that 15x15 analysis all else being equal.
Determine field center – The field size is measured along the center of the image in the inplane and crossplane direction. A 5mm strip is averaged and used to reduce noise.
Post-Analysis
Comparing centroids – The irradiated field centroid is compared to the EPID/image center as well as the the BB centroid. The field size is also reported.
IsoAlign#
New in version 3.15.
The IsoAlign phantom is for testing light/radiation coincidence. See also IsoAlign.
Image Acquisition#
The phantom should be placed on the couch at 100cm SSD.
Keep the phantom away from a couch edge or any rails.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom should be at a cardinal angle (0, 90, 180, or 270 degrees) relative to the EPID.
The phantom should be centered near the CAX (<2mm).
Analysis
Get BB centroid – An image window looks for the central BB as well as 1 BB in each cardinal direction. After finding the BBs, the centroid is calculated.
Determine field center – The field size is measured along the center of the image in the inplane and crossplane direction. A 10mm strip is averaged and used to reduce noise.
Post-Analysis
Comparing centroids – The irradiated field centroid is compared to the EPID/image center as well as the the BB centroid. The field size is also reported.
IMT L-Rad#
New in version 3.2.
The IMT L-Rad phantom is for testing light/radiation coincidence. Unlike the FC-2 phantom, the L-Rad’s BBs are all the way at the edge of the phantom. This means for any size below 20x20cm those BBs can’t be seen. Even at 20x20, the field edge partially obscures the BBs. For this reason, we only use the central BB for detection.
Image Acquisition#
The L-Rad phantom should be placed on the couch at 100cm SSD.
Keep the phantom away from a couch edge or any rails.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom should be at a cardinal angle (0, 90, 180, or 270 degrees) relative to the EPID.
The phantom should be centered near the CAX (<3mm).
Analysis
Get BB centroid – An image window looks for the central BB in a 1.2x1.2cm square. Once it finds it, the centroid is calculated.
Determine field center – The field size is measured along the center of the image in the inplane and crossplane direction. A 5mm strip is averaged and used to reduce noise.
Post-Analysis
Comparing centroids – The irradiated field centroid is compared to the EPID/image center as well as the the BB centroid. The field size is also reported.
SNC FSQA#
New in version 3.3.
The SNC FSQA phantom is for light/radiation coincidence. It contains markers which guide the physicist on how to position the light field for either a 10x10 or 15x15cm field. There is also an offset BB 4cm at the top right of the image. Because of both the philosophy of pylinac on light/rad and also because pylinac is a library and not a GUI, there is no interaction to find the edge markers. Instead, we use the one offset BB and then offset that point back 4cm in each direction to get a “virtual center”. This center is compared to the field center and EPID center. The expectation is that the physicist set up their field to the markers using the light field at the time of acquisition.
Image Acquisition#
The FSQA phantom should be placed on the couch at 100cm SSD.
Keep the phantom away from a couch edge or any rails.
Keep the phantom as close to 0 degrees rotation as possible.
Algorithm#
The algorithm works like such:
Allowances
The images can be acquired at any SID.
The images can be acquired with any EPID.
Restrictions
Warning
Analysis can fail or give unreliable results if any Restriction is violated.
The phantom should be at 0 degrees relative to the EPID.
The phantom should be roughly centered along the CAX (<3mm).
Analysis
Get BB centroid – An image window looks for the top-right offset BB in a 1.2x1.2cm square. Once it finds it, a “virtual center” centroid is calculated by shifting the detected BB location by 4cm in each direction.
Determine field center – The field size is measured along the center of the image in the inplane and crossplane direction. A 5mm strip is averaged and used to reduce noise.
Post-Analysis
Comparing centroids – The irradiated field centroid is compared to the EPID/image center as well as the the BB centroid. The field size is also reported.
Creating a custom phantom#
In the event you would like to analyze a phantom that pylinac does not analyze out of the box, the pylinac planar imaging module structure allows for generating new phantom analysis types quickly and easily. The benefit of this design is that with a few simple definitions you inherit a strong base of methods (e.g. plotting and PDF reports come for free).
Creating a new class involves a few different steps but can be done in a few minutes. The following is a guide for custom phantoms:
Subclass the
ImagePhantomBaseclass:
from pylinac.planar_imaging import ImagePhantomBase
class CustomPhantom(ImagePhantomBase):
pass
Define the
common_name. This is the name shown in plots and PDF reports.
class CustomPhantom(ImagePhantomBase):
common_name = "Custom Phantom v2.0"
If the phantom has a high-contrast measurement object, define the ROI locations.
class CustomPhantom(ImagePhantomBase):
...
high_contrast_roi_settings = {
"roi 1": {
"distance from center": 0.5,
"angle": 30,
"roi radius": 0.05,
"lp/mm": 0.2,
},
# add as many ROIs as are needed
}
Note
The exact values of your ROIs will need to be empirically determined. This usually involves an iterative process of adjusting the values until the values are satisfactory based on the ROI sample alignment to the actual ROIs.
If the phantom has a low-contrast measurement object, define the sample ROI and background ROI locations.
class CustomPhantom(ImagePhantomBase):
...
low_contrast_roi_settings = {
"roi 1": {
"distance from center": 0.5,
"angle": 30,
"roi radius": 0.05,
}, # no lp/mm key
# add as many ROIs as are needed
}
low_contrast_background_roi_settings = {
"roi 1": {"distance from center": 0.3, "angle": -45, "roi radius": 0.02},
# add as many ROIs as are needed
}
Note
The exact values of your ROIs will need to be empirically determined. This usually involves an iterative process of adjusting the values until the values are satisfactory based on the ROI sample alignment to the actual ROIs.
Set the “detection conditions”, which is the list of rules that must be true to properly detect the phantom ROI. E.g. the phantom should be near the center of the image. Detection conditions must always have a specific signature as shown below:
def my_special_detection_condition(
region: RegionProperties, instance: object, rtol: float
) -> bool:
# region is a scikit regionprop (https://scikit-image.org/docs/dev/api/skimage.measure.html#skimage.measure.regionprops)
# instance == self of the phantom
# rtol is relative tolerance of agreement. Don't have to use this.
do_stuff # e.g. is the region size and position correct?
return bool(result) # must always return a boolean
class CustomPhantom(ImagePhantomBase):
detection_conditions = [
my_special_detection_condition,
] # list of conditions; add as many as you want.
Optionally, add a phantom outline object. This helps visualize the algorithm’s determination of the size, center, and angle. If no object is defined, then no outline will be shown. This step is optional.
class CustomPhantom(ImagePhantomBase):
...
phantom_outline_object = {
"Circle": {"radius ratio": 0.5}
} # to create a circular outline
# or...
phantom_outline_object = {
"Rectangle": {"width ratio": 0.5, "height ratio": 0.3}
} # to create a rectangular outline
At this point you could technically call it done. You would need to always override the angle, center, and size values in the analyze method however. To automate this part you will need to fill in the associated logic. You can use whatever method you like. What I have found most useful is to use an edge detection algorithm and find the outline of the phantom.
class CustomPhantom(ImagePhantomBase):
...
def _phantom_center_calc(self) -> Point:
# do stuff in here to determine the center point location.
# don't forget to return as a Point item (pylinac.core.geometry.Point).
...
def _phantom_radius_calc(self) -> float:
# do stuff in here to return a float that represents the phantom radius value.
# This value does not have to relate to a physical measure. It simply defines a value that the ROIs scale by.
...
def _phantom_angle_calc(self) -> float:
# do stuff in here to return a float that represents the angle of the phantom.
# Again, this value does not have to correspond to reality; it simply offsets the ROIs.
# You may also return a constant if you like for any of these.
...
Congratulations! You now have a fully-functioning custom phantom. By using the base class and the predefined attributes and methods, the plotting and PDF report functionality comes for free.
Usage tips, tweaks, & troubleshooting#
Set the SSD of your phantom#
If your phantom is at a non-standard distance (!= 1000mm), e.g. sitting on the EPID panel, you can specify its
distance via the ssd parameter.
Warning
The ssd should be in mm, not cm. Pylinac is moving toward consistent units on everything and it will be mm for distance.
from pylinac import StandardImagingQC3
qc = StandardImagingQC3(...)
qc.analyze(..., ssd=1500) # distance to the phantom in mm.
Adjust an ROI on an existing phantom#
To adjust an ROI, override the relevant attribute or create a subclass. E.g. to move the 2nd ROI of the high-contrast ROI set of the QC-3 phantom:
from pylinac import StandardImagingQC3
StandardImagingQC3.high_contrast_roi_settings["roi 1"][
"distance from center"
] = 0.05 # overrides that one setting
qc3 = StandardImagingQC3(...)
# or
class TweakedStandardImagingQC3(StandardImagingQC3):
high_contrast_roi_settings = {
"roi 1": ...
} # note that you must replace ALL the values
qc3 = TweakedStandardImagingQC3(...)
Calculating Uniformity#
The uniformity of the phantom can be found by using the percent_integral_uniformity() method.
This uses the same equation as ACR for CT uniformity. See the “Uniformity” section under ACR Analysis for more information.
The PIU is calculated over all the low-contrast ROIs and the lowest (worst) PIU is returned.
For robustness, the 1st and 99th percentiles are used rather than the min/max. The true min/max can be influenced by salt and paper noise. To use the true min and max, set the percentiles to 0 and 100 respectively:
Note
This will not be reflected in the results_data structure.
leeds = LeedsTOR(...)
leeds.analyze(...)
print(leeds.percent_integral_uniformity(percentiles=(0, 100))) # uses the true min/max
Warning
This equation was chosen because it is common and understood, but it does come with pitfalls. It is not designed to handle negative values or 0. The calculated result may be misleading if these conditions exist.
Calculate a specific MTF#
To calculate a specific MTF value, i.e. the frequency at a given MTF%:
dl = DoselabMC2kV(...)
dl.analyze(...)
print(dl.mtf.relative_resolution(x=50)) # 50% rMTF
Get/View the contrast of a low-contrast ROI#
leeds = LeedsTOR(...)
leeds.analyze(...)
print(leeds.low_contrast_rois[1].contrast) # get the 2nd ROI contrast value
Loosen the ROI finding conditions#
If for some reason you have a need to loosen the existing phantom-finding algorithm conditions you can do so fairly easily by overloading the current tooling:
from pylinac.planar_imaging import is_right_size, is_centered, LeedsTOR
def is_right_size_loose(region, instance, rtol=0.3): # rtol default is 0.1
return is_right_size(region, instance, rtol)
# set the new condition for whatever
LeedsTOR.detection_conditions = [is_right_size_loose, is_centered]
# proceed as normal
myleeds = LeedsTOR(...)
Scaling#
New in version 3.19.
Pylinac can produce an area calculation of the phantom. This can be used as a way to test the scaling of the imager per TG-142. The scaling is based on the blue rectangle/circle that is shown in the plots.
E.g.:
leeds = pylinac.LeedsTOR(...)
leeds.analyze(...)
results = leeds.results_data()
print(results.phantom_area) # in mm^2
Warning
The produced scaling value is based on the blue rectangle/circle. In many cases it does not equal the exact size of the phantom. It is recommended to be used as a constancy check.
Adjusting the scaling#
If you are dead-set on having the scaling value be the exact size of the phantom,
or you simply have a different interpretation of what the scaling should be you
can override the scaling calculation to a degree. The scaling is calculated
using the phantom_outline_object attribute. This attribute is a dictionary
and defines the size of the rectangle/circle that is shown in the plots. Changing
these values will both change the plot and the area/scaling value.
import pylinac
class NewSNCkV(pylinac.SNCkV):
phantom_outline_object = {
"Rectangle": {"width ratio": 8.4, "height ratio": 7.2} # change these
}
class NewLeeds(pylinac.LeedsTOR):
phantom_outline_object = {"Circle": {"radius ratio": 1.3}} # change this
Wrong phantom angle#
It may sometimes be that the angle of the phantom appears incorrect, or the results appear incorrect. E.g. here is a QC-3 phantom:
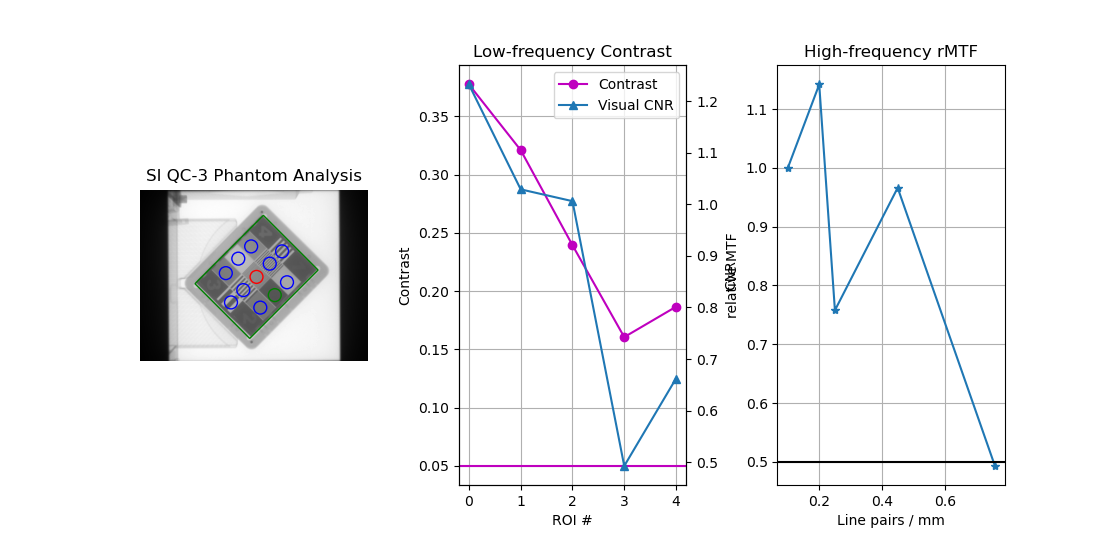
The ROIs appear correct, the but the contrast and MTF do not monotonically decrease, indicating a problem. In this case, it is because the image acquisition rules were not followed. For the QC-3, the “1” should always point toward the gantry, as per the manual. When oriented this way, the results will be correct.
Light/Radiation philosophy#
Pylinac (or rather the author) has an opinionated philosophy about light vs radiation that affects the way light/radiation analysis is performed. In our opinion, light/rad using a phantom is antiquated as EPIDs are robust enough nowadays to be quite reliable, at least for Varian machines. By using something as simple as graph paper after mechanical measurements, a light field can be set and a simple open field delivered. This open field size and CAX offset can be compared to the nominal values set by the physicist at the time of acquisition.
Short of using CCD cameras or specialty equipment like phosphorus, there is no true way to know the light field measurement. All we have is what the physicist set up to. If the physicist sets up to a nominal size like 10x10, then a radiation field measurement can be compared to that rather simply with common field analysis. E.g if the measured field size was 10.1x10.6mm then the error between light and rad is 0.1 and 0.6mm respectively. The CAX offset follows the same logic.
You may disagree, but this is here for the purposes of explaining our philosophy and why light/rad does (or does not do) what it does.
We provide these light/rad routines because customers ask for them, not because we recommend them.
API Documentation#
- class pylinac.planar_imaging.LeedsTOR(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.LeedsTORBlue(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
LeedsTORThe Leeds TOR (Blue) is for analyzing older Leeds phantoms which have slightly offset ROIs compared to the newer, red-ring variant.
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- static run_demo() None#
Run the Leeds TOR phantom analysis demonstration.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.StandardImagingQC3(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.StandardImagingQCkV(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
StandardImagingQC3Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.LasVegas(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- results(as_list: bool = False) str | list[str][source]#
Return the results of the analysis. Overridden because ROIs seen is based on visibility, not CNR.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.ElektaLasVegas(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
LasVegasElekta’s variant of the Las Vegas.
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis. Overridden because ROIs seen is based on visibility, not CNR.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.DoselabMC2MV(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
DoselabMC2kVParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.DoselabMC2kV(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.SNCMV(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
SNCkVParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.SNCMV12510(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
SNCMVThe older SNC MV QA phantom w/ model number 1251000
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- static run_demo() None#
Run the Sun Nuclear MV-QA phantom analysis demonstration.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.SNCkV(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.PTWEPIDQC(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.IBAPrimusA(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- window_floor() float | None[source]#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- window_ceiling() float | None[source]#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- property phantom_angle: float#
Cache this; calculating the angle is expensive
- analyze(low_contrast_threshold: float = 0.05, high_contrast_threshold: float = 0.5, invert: bool = False, angle_override: float | None = None, center_override: tuple | None = None, size_override: float | None = None, ssd: float | Literal['auto'] = 'auto', low_contrast_method: str = 'Michelson', visibility_threshold: float = 100) None#
Analyze the phantom using the provided thresholds and settings.
Parameters#
- low_contrast_thresholdfloat
This is the contrast threshold value which defines any low-contrast ROI as passing or failing.
- high_contrast_thresholdfloat
This is the contrast threshold value which defines any high-contrast ROI as passing or failing.
- invertbool
Whether to force an inversion of the image. This is useful if pylinac’s automatic inversion algorithm fails to properly invert the image.
- angle_overrideNone, float
A manual override of the angle of the phantom. If None, pylinac will automatically determine the angle. If a value is passed, this value will override the automatic detection.
Note
0 is pointing from the center toward the right and positive values go counterclockwise.
- center_overrideNone, 2-element tuple
A manual override of the center point of the phantom. If None, pylinac will automatically determine the center. If a value is passed, this value will override the automatic detection. Format is (x, y)/(col, row).
- size_overrideNone, float
A manual override of the relative size of the phantom. This size value is used to scale the positions of the ROIs from the center. If None, pylinac will automatically determine the size. If a value is passed, this value will override the automatic sizing.
Note
This value is not necessarily the physical size of the phantom. It is an arbitrary value.
- ssd
The SSD of the phantom itself in mm. If set to “auto”, will first search for the phantom at the SAD, then at 5cm above the SID.
- low_contrast_method
The equation to use for calculating low contrast.
- visibility_threshold
The threshold for whether an ROI is “seen”.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(image: bool = True, low_contrast: bool = True, high_contrast: bool = True, show: bool = True, split_plots: bool = False, **plt_kwargs: dict) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- imagebool
Show the image.
- low_contrastbool
Show the low contrast values plot.
- high_contrastbool
Show the high contrast values plot.
- showbool
Whether to actually show the image when called.
- split_plotsbool
Whether to split the resulting image into individual plots. Useful for saving images into individual files.
- plt_kwargsdict
Keyword args passed to the plt.figure() method. Allows one to set things like figure size.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
Parameters#
- as_listbool
Whether to return as a list of strings vs single string. Pretty much for internal usage.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, split_plots: bool = False, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- split_plots: bool
If split_plots is True, multiple files will be created that append a name. E.g. my_file.png will become my_file_image.png, my_file_mtf.png, etc. If to_streams is False, a list of new filenames will be returned
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- class pylinac.planar_imaging.StandardImagingFC2(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
ImagePhantomBaseParameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(invert: bool = False, fwxm: int = 50, bb_edge_threshold_mm: float = 10) None[source]#
Analyze the FC-2 phantom to find the BBs and the open field and compare to each other as well as the EPID.
Parameters#
- invertbool
Whether to force-invert the image from the auto-detected inversion.
- fwxmint
The FWXM value to use to detect the field. For flattened fields, the default of 50 should be fine. For FFF fields, consider using a lower value such as 25-30.
- bb_edge_threshold_mmfloat
The threshold in mm to use to determine if the BB is near the edge of the image. If the BB is within this threshold, a different algorithm is used to determine the BB position that is more robust to edge effects but can give uncertainty when in a flat region (i.e. away from the field edge).
- plot_analyzed_image(show: bool = True, **kwargs) tuple[list[Figure], list[str]][source]#
Plot the analyzed image.
Parameters#
- showbool
Whether to actually show the image when called.
- save_analyzed_image(filename: None | str | BinaryIO = None, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None[source]#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)[source]#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.IMTLRad(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
StandardImagingFC2The IMT light/rad phantom: https://www.imtqa.com/products/l-rad
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(invert: bool = False, fwxm: int = 50, bb_edge_threshold_mm: float = 10) None#
Analyze the FC-2 phantom to find the BBs and the open field and compare to each other as well as the EPID.
Parameters#
- invertbool
Whether to force-invert the image from the auto-detected inversion.
- fwxmint
The FWXM value to use to detect the field. For flattened fields, the default of 50 should be fine. For FFF fields, consider using a lower value such as 25-30.
- bb_edge_threshold_mmfloat
The threshold in mm to use to determine if the BB is near the edge of the image. If the BB is within this threshold, a different algorithm is used to determine the BB position that is more robust to edge effects but can give uncertainty when in a flat region (i.e. away from the field edge).
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(show: bool = True, **kwargs) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- showbool
Whether to actually show the image when called.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- static run_demo() None#
Run the Standard Imaging FC-2 phantom analysis demonstration.
- save_analyzed_image(filename: None | str | BinaryIO = None, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.DoselabRLf(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
StandardImagingFC2The Doselab light/rad phantom
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(invert: bool = False, fwxm: int = 50, bb_edge_threshold_mm: float = 10) None#
Analyze the FC-2 phantom to find the BBs and the open field and compare to each other as well as the EPID.
Parameters#
- invertbool
Whether to force-invert the image from the auto-detected inversion.
- fwxmint
The FWXM value to use to detect the field. For flattened fields, the default of 50 should be fine. For FFF fields, consider using a lower value such as 25-30.
- bb_edge_threshold_mmfloat
The threshold in mm to use to determine if the BB is near the edge of the image. If the BB is within this threshold, a different algorithm is used to determine the BB position that is more robust to edge effects but can give uncertainty when in a flat region (i.e. away from the field edge).
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(show: bool = True, **kwargs) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- showbool
Whether to actually show the image when called.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.IsoAlign(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
StandardImagingFC2The PTW Iso-Align light/rad phantom
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(invert: bool = False, fwxm: int = 50, bb_edge_threshold_mm: float = 10) None#
Analyze the FC-2 phantom to find the BBs and the open field and compare to each other as well as the EPID.
Parameters#
- invertbool
Whether to force-invert the image from the auto-detected inversion.
- fwxmint
The FWXM value to use to detect the field. For flattened fields, the default of 50 should be fine. For FFF fields, consider using a lower value such as 25-30.
- bb_edge_threshold_mmfloat
The threshold in mm to use to determine if the BB is near the edge of the image. If the BB is within this threshold, a different algorithm is used to determine the BB position that is more robust to edge effects but can give uncertainty when in a flat region (i.e. away from the field edge).
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(show: bool = True, **kwargs) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- showbool
Whether to actually show the image when called.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- save_analyzed_image(filename: None | str | BinaryIO = None, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background
- class pylinac.planar_imaging.SNCFSQA(filepath: str | BinaryIO | Path, normalize: bool = True, image_kwargs: dict | None = None)[source]#
Bases:
StandardImagingFC2SNC light/rad phantom. See the ‘FSQA’ phantom and specs: https://www.sunnuclear.com/products/suncheck-machine.
Unlike other light/rad phantoms, this does not have at least a centered BB. The edge markers are in the penumbra and thus detecting them is difficult. We thus detect the one offset marker in the top right of the image. This is offset by 4cm in each direction. We can then assume that the phantom center is -4cm from this point, creating a ‘virtual center’ so we have an apples-to-apples comparison.
Parameters#
- filepathstr
Path to the image file.
- normalize: bool
Whether to “ground” and normalize the image. This can affect contrast measurements, but for backwards compatibility this is True. You may want to set this to False if trying to compare with other software.
- image_kwargsdict
Keywords passed to the image load function; this would include things like DPI or SID if applicable
- analyze(invert: bool = False, fwxm: int = 50, bb_edge_threshold_mm: float = 10) None#
Analyze the FC-2 phantom to find the BBs and the open field and compare to each other as well as the EPID.
Parameters#
- invertbool
Whether to force-invert the image from the auto-detected inversion.
- fwxmint
The FWXM value to use to detect the field. For flattened fields, the default of 50 should be fine. For FFF fields, consider using a lower value such as 25-30.
- bb_edge_threshold_mmfloat
The threshold in mm to use to determine if the BB is near the edge of the image. If the BB is within this threshold, a different algorithm is used to determine the BB position that is more robust to edge effects but can give uncertainty when in a flat region (i.e. away from the field edge).
- classmethod from_demo_image()#
Instantiate and load the demo image.
- property magnification_factor: float#
The mag factor of the image based on SSD vs SAD
- percent_integral_uniformity(percentiles: tuple[float, float] = (1, 99)) float | None#
Calculate and return the percent integral uniformity (PIU). This uses a similar equation as ACR does for CT protocols. The PIU is calculated over all the low contrast ROIs and the lowest (worst) PIU is returned.
If the phantom does not contain low-contrast ROIs, None is returned.
- property phantom_area: float#
The area of the detected ROI in mm^2
- property phantom_bbox_size_px: float#
The phantom bounding box size in pixels^2 at the isoplane.
- property phantom_ski_region: RegionProperties#
The skimage region of the phantom outline.
- plot_analyzed_image(show: bool = True, **kwargs) tuple[list[Figure], list[str]]#
Plot the analyzed image.
Parameters#
- showbool
Whether to actually show the image when called.
- publish_pdf(filename: str, notes: str = None, open_file: bool = False, metadata: dict | None = None, logo: Path | str | None = None)#
Publish (print) a PDF containing the analysis, images, and quantitative results.
Parameters#
- filename(str, file-like object}
The file to write the results to.
- notesstr, list of strings
Text; if str, prints single line. If list of strings, each list item is printed on its own line.
- open_filebool
Whether to open the file using the default program after creation.
- metadatadict
Extra data to be passed and shown in the PDF. The key and value will be shown with a colon. E.g. passing {‘Author’: ‘James’, ‘Unit’: ‘TrueBeam’} would result in text in the PDF like: ————– Author: James Unit: TrueBeam ————–
- logo: Path, str
A custom logo to use in the PDF report. If nothing is passed, the default pylinac logo is used.
- results(as_list: bool = False) str | list[str]#
Return the results of the analysis.
- results_data(as_dict: bool = False, as_json: bool = False) T | dict | str#
Present the results data and metadata as a dataclass, dict, or tuple. The default return type is a dataclass.
Parameters#
- as_dictbool
If True, return the results as a dictionary.
- as_jsonbool
If True, return the results as a JSON string. Cannot be True if as_dict is True.
- static run_demo() None#
Run the Standard Imaging FC-2 phantom analysis demonstration.
- save_analyzed_image(filename: None | str | BinaryIO = None, to_streams: bool = False, **kwargs) dict[str, BinaryIO] | list[str] | None#
Save the analyzed image to disk or to stream. Kwargs are passed to plt.savefig()
Parameters#
- filenameNone, str, stream
A string representing where to save the file to. If split_plots and to_streams are both true, leave as None as newly-created streams are returned.
- to_streams: bool
This only matters if split_plots is True. If both of these are true, multiple streams will be created and returned as a dict.
- window_ceiling() float | None#
The value to use as the maximum when displaying the image. Helps show contrast of images, specifically if there is an open background
- window_floor() float | None#
The value to use as the minimum when displaying the image (see https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.imshow.html) Helps show contrast of images, specifically if there is an open background